Genomic Sequencing of Ancient DNA Illuminates Plague Origins
The Black Plague (1346-1353) is one of the most widely recognized historic outbreaks of pandemic disease. It was caused by the Yersinia pestis bacterium, which is generally thought to have spread to humans through fleas living on small rodents, a transmission model extrapolated from the scientific understanding of modern Y. pestis transmission and disease manifestation. To date, there have been 3 DNA-verified Y. pestis pandemics (the Justinian Plague, Black Death and Modern Plague) during recorded history. However, ancient DNA suggests that these are not the only instances of the organism plaguing humans, and data indicate that additional vectors have likely played a role in disease transmission throughout history.
Identification of Y. pestis
At the end of the 18th century, when a bubonic disease emerged again, it achieved global distribution. The reemergence coincided with widespread acceptance of germ theory and primed scientists the world over for the discovery of Y. pestis.
In 1894 this familiar, but unidentified, disease was spreading throughout Hong Kong, and a number of scientists converged on the city to try to identify the causative agent of the "Hong Kong epidemic." Using Koch’s postulates and light microscopy, Alexadre Yersin and Shibasaburo Kitasato independently described Y. pestis as the microbiological cause of the epidemic, and Alexadre Yersin linked rats to the transmission cycle soon after.
A year later, Masanori Ogata and Paul-Louis Simond simultaneously identified black rat fleas, Xenopsylla cheopsis, as the primary vector of transmission between black rats and humans. Yersin then hypothesized that Y. pestis was the cause of many historic plagues, including the Black Death and the Justinian Plague, which had similar written descriptions. However, other hypotheses of the causative agent remained and were hard to dispute until molecular approaches, including PCR, conclusively linked Y. pestis to the Black Plague.
With More Data Come More Questions
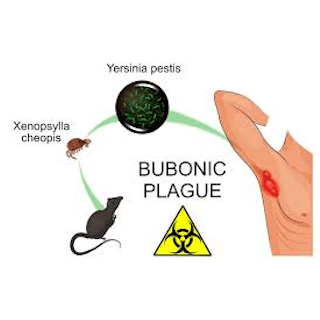
As with countless other subfields of microbiology, next-generation sequencing caused seismic shifts in our understanding of Y. pestis and highlighted a number of discrepancies that encouraged further study. Modern Y. pestis contains the gene ymt which allows it to be stably maintained in the flea gut and transmitted to the human host via biofilm-dependent transmission. In this method of transmission, Y. pestis creates a biofilm that blocks normal function in the flea’s gut and impairs its ability to feed, leading to more regular flea biting and mixing of the bacteria with host blood at the bite wound. Unexpectedly, sequenced genomes of Y. pestis collected from skeletal remains from some ancient plague victims lack the ymt gene.
Paleomicrobiologists have isolated Y. pestis genomic DNA from dental pulp as much as 5,000 years old, indicating that Y. pestis has been associated with humans for much longer than previously expected. The lack of ymt leaves an open question about the origin and degree of virulence of early Y. pestis and suggests that additional vectors may have been involved in the spread of Y. pestis in ancient populations. Other proposed vectors include Pediculus humanus, the human body louse, and Pulex irritans, the human flea. These organisms both live in close association with humans, are known to carry Y. pestis during plague epidemics and pass viable Y. pestis in their feces that can be scratched into the skin. The transmission patterns of these vectors also more closely mirror those observed during the second pandemic.
Furthermore, the black rat is not thought to have been broadly distributed in Europe during the Middle Ages because of cold temperatures associated with "The Little Ice Age," making it unlikely that X. cheopsis would have transmitted Y. pestis. Fossils of X. cheopsis have not been found in Europe concurrent with the second plague pandemic, but P. irritansfossils have been found. These findings suggest that P. irritans is a flea more likely responsible for transmission of the Black Plague than X. cheopsis.
Additionally, most clinical samples of Y. pestis are nearly clonal, which suggests a single common ancestor and low variability in natural populations. However, ancient samples indicate that high diversity was once present. While some Bronze age isolates lack ymt, the ymt gene has been detected in ancient DNA samples recovered from Early Bronze Age Russian remains, suggesting that multiple strains may have circulated concurrently. This ymt gene variability suggests that different regions may have experienced different primary modes of transmission and perhaps even slight variation in disease manifestation.
Finally, despite the importance of other genes to Y. pestis infection, ymt has historically garnered the most study in sequencing, so it is hard to predict the full impact of strain diversity on historic human disease outcomes. Nucleotide variations observed in the genomes of ancient strains from Neolithic and Bronze age burials indicate the presence of diversification events that led to new strains that spread to different regions. This evidence suggests that, over time, many strains went extinct while others evolved into the present-day strain. Mapping these variations has helped to generate hypotheses about routes of global distribution.
Outstanding Questions and Future Work
Next-generation sequencing of ancient DNA samples collected from plague victims has advanced the scientific understanding of the origin and transmission of one of the most notorious infectious diseases known to humankind. Despite these advances in knowledge, many questions surrounding Y. pestis still remain. Due to limited sequencing for Y. pestis outside of Europe, there is incomplete understanding of how genotypic changes might be associated with, or impacted by, latent periods between outbreaks and reemergence of disease for each pandemic period. More robust sequencing in Africa and Asia will enable scientists to begin filling in these timelines. Additional work is focused on uncovering the role of soil-based reservoirs of Y. pestis, including amoeba that may harbor the bacteria during the quiescent periods between outbreaks. Answering these questions will help inform our understanding of the downtime between waves of infections and prepare us for future pandemics.
