Whole Genome Sequencing (WGS) as a Tool for Hospital Surveillance
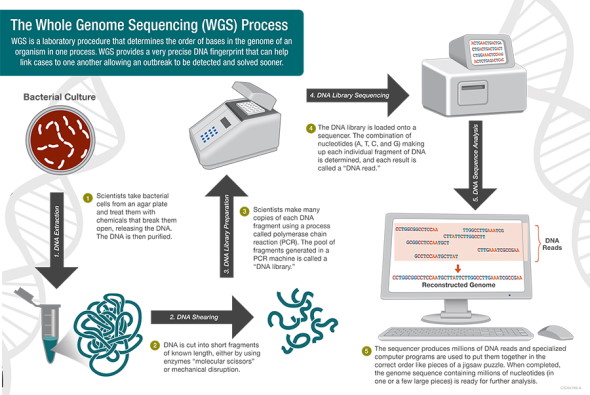
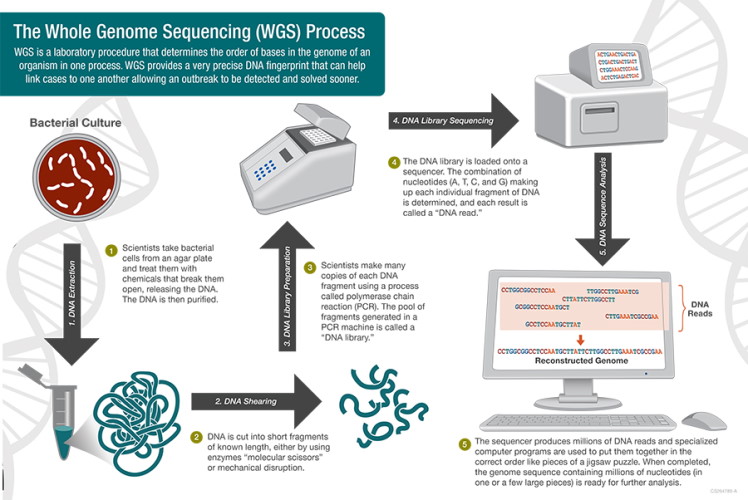
At the most basic level, genome sequencing is the science of "reading" the order of the base pairs (adenine, thymine, cytosine and guanine) that make up an organism's DNA. Once this order is read, a complete genetic picture of the organism is formed, akin to a unique fingerprint. Historically, this technology has been beneficial for identifying outbreaks and performing public health surveillance. However, the benefits and challenges of implementation of whole genome sequencing (WGS) in the hospital setting are less clear.
Whole Genome Sequencing in Outbreak Investigation and Surveillance
The term whole genome sequencing may bring to mind the revolutionary Human Genome Project, which involved thousands of scientists and led to the complete sequencing of the entire human genome. While WGS of bacteria is much simpler than of humans, it is highly impactful. A shining example of WGS put to work in the real world is the Centers for Disease Control and Prevention's (CDC) PulseNet national laboratory network. Until recently, the PulseNet network used pulse-field gel electrophoresis (PFGE) to identify the unique fingerprints (pattern designation) of bacteria that cause foodborne illness and stamp out outbreaks across the U.S. This methodology has prevented over 200,000 illnesses and saved nearly half a billion dollars in medical costs and missed school or work.
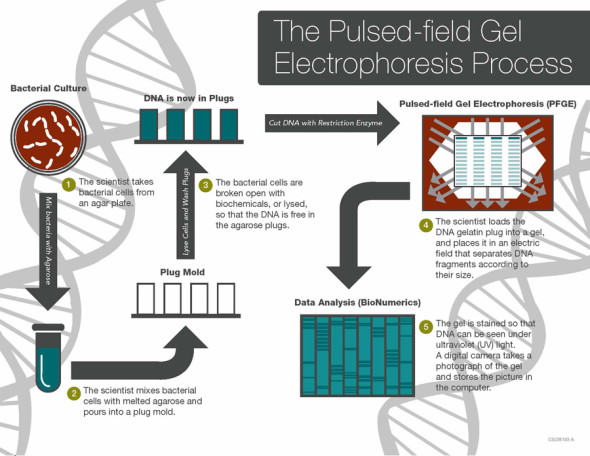
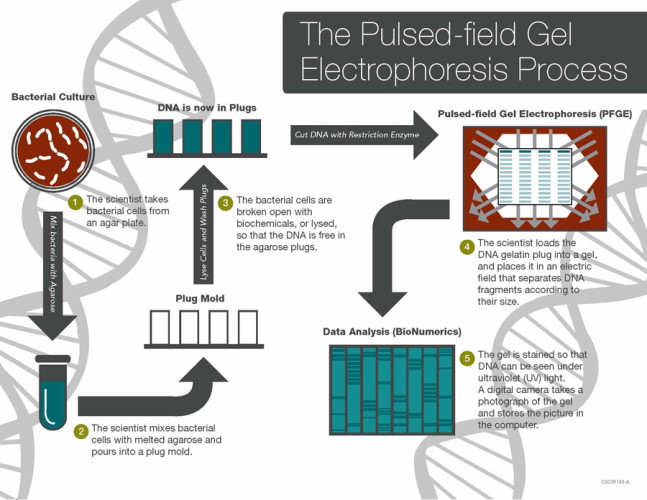
In 2012, the Food and Drug Administration (FDA) began using WGS regularly to monitor and protect the national food supply. In 2019, PulseNet (CDC) made the transition from PFGE to WGS. Compared to PFGE, WGS provides more detailed and precise data, ultimately allowing scientists to compare millions of base pairs within the bacterial DNA. As the CDC describes it, WGS "is like comparing all of the words in a book (WGS), instead of just the number of chapters (PFGE), to see if the books are the same or different." The advantage of having more detailed genomic data is that the source of outbreaks can be determined sooner and with greater precision, since the exact details of the organism and where it came from can be determined more effectively with WGS. In addition, for some organizations like PulseNet, the technology will improve surveillance efficiency when combined with epidemiologic work and bioinformatics. The CDC's sequencing of Listeria monocytogenes began in 2013 and laid the groundwork for sequencing of other foodborne pathogens that has now become standard.
A timely example of WGS in public health is the CDC's June 30, 2021 Health Alert Network message, detailing the infection of 3 individuals with Burkholderia pseudomallei, which causes the disease melioidosis, who have no documented travel history outside of the U.S. Based on genomic analysis, epidemiologists at the CDC were able to determine that all 3 cases (found geographically dispersed in Kansas, Texas and Minnesota) likely share a common source. The source of the infections has not yet been confirmed.
Benefits of WGS for Hospital Surveillance and Infection Prevention
The precise and detailed data that WGS provides can be especially beneficial to infection prevention efforts in the hospital setting. For example, WGS can be used to identify the environmental source of an outbreak, trace the transmission of infectious agents between patients and better understand the transmission dynamics of antimicrobial resistance genes. Historically, traditional microbiological methods, such as susceptibility pattern comparison and PFGE, have been used to identify and prevent hospital-acquired infections and associated outbreaks. However, most of these methods cannot truly determine organism relatedness. Since PFGE examines large sections of genetic information, minute genetic details that differentiate organisms or determine relatedness may be missed. The same is also true for susceptibility patterns, which may differ over time in the same organism, or look very similar between 2 genetically different organisms. Additionally, changes in organism susceptibility patterns are often due to the loss or gain of mobile genetic elements harboring resistance. In addition to providing more precise data, hospital-based WGS can provide data faster during an acute outbreak.
A recent study demonstrated that implementing WGS into the infection-prevention workflow can significantly impact patient mortality and hospital costs. In this study, the standard of care was defined as the use of WGS only during an outbreak. The proposed intervention was to perform WGS on all clinical isolates of selected pathogens that are known to cause severe disease, are associated with antimicrobial resistance or have been known to cause hospital-associated outbreaks, regardless of the presence of an outbreak. The investigator's primary hypothesis was that using WGS as a tool of regular surveillance would detect outbreaks that the standard of care method would not otherwise detect.
During the study period, there were 11 outbreaks that included 89 patients. The cost-effectiveness analysis demonstrated that if WGS surveillance had been in place during the study period, 46% of transmissions could have been avoided, and nearly $45,000 would have been saved, despite the cost of WGS testing. Overall, the results of this study suggest that implementing WGS as a regular infection prevention surveillance tool is more favorable than the standard of care practice of only performing WGS when an outbreak has been identified.
However, there are some critical caveats: in this study, a WGS surveillance-based infection prevention program became less favorable when the proportion of patients with positive respiratory cultures increased (colonization with normal microbiota was often treated with antibiotics and cost-savings in infection treatment decreased), when culture results were delayed and when the response or intervention by infection preventionists was delayed. This is important to note because while the technology is useful, the benefits become nullified if there is not an actionable and timely response to the WGS results. These caveats highlight the importance of infrastructure for hospital-based WGS surveillance, one of the primary challenges associated with implementation.
Barriers and Challenges to Implementing WGS in the Clinical Microbiology Laboratory
WGS technology has advanced quickly, and it is now reasonable for clinical microbiology laboratories to consider implementing WGS in-house without sending isolates to central laboratories or public health departments. However, this option is not without significant barriers that must be considered. As is true of any stewardship or infection-prevention initiative, advanced technologies lose their potential impact when a solid infrastructure that supports the testing is not in place. Some key barriers to WGS surveillance and implementation in the clinical microbiology laboratory include the following:
Cost
The cost associated with testing platforms, consumables and training or hiring of testing staff and staff with bioinformatics experience is high. For laboratories that are not currently performing genotyping or PFGE (tests that WGS would ultimately replace), a cost-benefit analysis will likely be necessary to convince hospital administrators that such an implementation is beneficial and worth the cost.
Expertise
WGS implementation requires skilled technicians for the "wet-lab" component of the testing, which involves extraction, library preparation and sequencing, and "dry-lab" expertise, which involves bioinformatics. While the technology may be relatively easy to use, interpreting the large amounts of data produced by WGS requires highly trained staff with a specific skill set.
Information Technology Infrastructure
One vital component that is often overlooked is the information technology (IT) infrastructure needed to implement WGS. Seemingly simple things, like internet speed and data storage, are imperative to ensuring large amounts of sequencing data can be processed.
Data Sharing and Communication
Communication between the microbiology laboratory, infection preventionists and infectious disease clinicians is crucial when implementing new technologies in the laboratory. If WGS surveillance is to have a real-world impact on outbreak prevention and management, the data gleaned from sequencing must be communicated to infection prevention professionals in a clear and timely manner. As described above, WGS surveillance implementations become less favorable when infection preventionists are delayed in their outbreak intervention. Additionally, genomic data are best interpreted when they can be compared with other genomic sequences. Sharing WGS findings with a local public health department is also critical, as they are often well-equipped to assist in outbreak investigation and the interpretation of WGS results. Therefore, it is essential to consider a hospital's ability to share microbiologic, patient and infection control data with other institutions.
Quality Control and Standardization
Quality control is at the core of the clinical microbiology laboratory, and it is crucial for interpreting all tests performed therein. Currently, standardized quality control measures for WGS have not been established, making test implementation and interpretation challenging. In addition, this lack of national and international standardization may pose unique challenges to data sharing if tests are performed and analyzed differently.
Overall, WGS demonstrates a tremendous amount of promise as a tool for hospital-based surveillance. While the technology has developed to the point that it may be feasible for microbiology laboratories to implement, developing a supportive infrastructure within the hospital requires a significant amount of thought and investment. Laboratories considering the implementation of WGS for surveillance should make every effort to ensure that the testing fits within the clinical microbiology workflow and that WGS results are interpretable and actionable, with emphasis placed on clear communication between the microbiology laboratory and infection preventionists.
